MERFISH and scRNA data preprocess
We apply uniPort to integrate high-plex RNA imaging-based spatially resolved MERFISH data with scRNA-seq data. The MERFISH data includes 64,373 cells with 155 genes, and the scRNA-seq data includes 30,370 cells with 21,030 genes from six mice measured with dissociated scRNA-seq (10X).
[1]:
import uniport as up
import scanpy as sc
import pandas as pd
import numpy as np
up.__version__
[1]:
'1.1.1'
Read the cell type annotations of MERFISH and scRNA-seq data seperately.
[2]:
labels_merfish = pd.read_csv('MERFISH/MERFISH_st_filter_cluster.txt', sep='\t')
celltype_merfish = labels_merfish['cluster_main'].values
labels_rna = pd.read_csv('MERFISH/MERFISH_scRNA_filter_cluster.txt', sep='\t')
celltype_rna = labels_rna['cluster_main'].values
Read MERFISH and scRNA-seq into AnnData objects using load_file fucntion in uniport.
[3]:
# adata_merfish = sc.read_h5ad('MERFISH/merfish0.h5ad')
# adata_rna = sc.read_h5ad('MERFISH/rna0.h5ad')
adata_merfish = up.load_file('MERFISH/MERFISH_mouse1.txt')
adata_rna = up.load_file('MERFISH/RNA_count.txt')
AnnDataobjects.[4]:
adata_merfish.obs['cell_type'] = celltype_merfish
adata_merfish.obs['domain_id'] = 0
adata_merfish.obs['domain_id'] = adata_merfish.obs['domain_id'].astype('category')
adata_merfish.obs['source'] = 'MERFISH'
adata_rna.obs['cell_type'] = celltype_rna
adata_rna.obs['domain_id'] = 1
adata_rna.obs['domain_id'] = adata_rna.obs['domain_id'].astype('category')
adata_rna.obs['source'] = 'RNA'
print(adata_rna.obs)
print(adata_merfish.obs)
cell_type domain_id source
AAACCTGAGATGTGGC-1 Fibroblast 1 RNA
AAACCTGCACACAGAG-1 Excitatory 1 RNA
AAACCTGCACTACAGT-1 Inhibitory 1 RNA
AAACCTGTCAGGATCT-1 Excitatory 1 RNA
AAACCTGTCGCACTCT-1 OD Mature 1 RNA
... ... ... ...
TTTGGTTGTTATCACG-6 Inhibitory 1 RNA
TTTGGTTGTTATTCTC-6 Inhibitory 1 RNA
TTTGTCAGTTCCGTCT-6 Inhibitory 1 RNA
TTTGTCATCGTGGGAA-6 Inhibitory 1 RNA
TTTGTCATCTTTACAC-6 Excitatory 1 RNA
[30370 rows x 3 columns]
cell_type domain_id source
cell1 Astrocyte 0 MERFISH
cell2 Inhibitory 0 MERFISH
cell3 Inhibitory 0 MERFISH
cell4 Inhibitory 0 MERFISH
cell5 Inhibitory 0 MERFISH
... ... ... ...
cell73650 OD Mature 0 MERFISH
cell73651 OD Mature 0 MERFISH
cell73653 OD Immature 0 MERFISH
cell73654 OD Mature 0 MERFISH
cell73655 OD Mature 0 MERFISH
[64373 rows x 3 columns]
Concatenate scATAC-seq and scRNA-seq with common genes using AnnData.concatenate.
[5]:
adata_cm = adata_merfish.concatenate(adata_rna, join='inner', batch_key='domain_id')
print(adata_cm.obs)
cell_type domain_id source
cell1-0 Astrocyte 0 MERFISH
cell2-0 Inhibitory 0 MERFISH
cell3-0 Inhibitory 0 MERFISH
cell4-0 Inhibitory 0 MERFISH
cell5-0 Inhibitory 0 MERFISH
... ... ... ...
TTTGGTTGTTATCACG-6-1 Inhibitory 1 RNA
TTTGGTTGTTATTCTC-6-1 Inhibitory 1 RNA
TTTGTCAGTTCCGTCT-6-1 Inhibitory 1 RNA
TTTGTCATCGTGGGAA-6-1 Inhibitory 1 RNA
TTTGTCATCTTTACAC-6-1 Excitatory 1 RNA
[94743 rows x 3 columns]
Preprocess data using functions normalize_total, log1p and highly_variable_genes in scanpy and batch_scale in uniport (modified from SCALEX)
[6]:
sc.pp.normalize_total(adata_cm)
sc.pp.log1p(adata_cm)
sc.pp.highly_variable_genes(adata_cm, n_top_genes=2000, batch_key='domain_id', inplace=False, subset=True)
up.batch_scale(adata_cm)
... storing 'cell_type' as categorical
... storing 'source' as categorical
[7]:
sc.pp.normalize_total(adata_merfish)
sc.pp.log1p(adata_merfish)
sc.pp.highly_variable_genes(adata_merfish, n_top_genes=2000, inplace=False, subset=True)
up.batch_scale(adata_merfish)
[8]:
sc.pp.normalize_total(adata_rna)
sc.pp.log1p(adata_rna)
sc.pp.highly_variable_genes(adata_rna, n_top_genes=2000, inplace=False, subset=True)
up.batch_scale(adata_rna)
Save the preprocessed data for integration and online prediction.
[9]:
adata_merfish.write('MERFISH/MERFISH_processed.h5ad', compression='gzip')
adata_rna.write('MERFISH/RNA_processed.h5ad', compression='gzip')
adata_cm.write('MERFISH/MERFISH_and_RNA.h5ad', compression='gzip')
... storing 'cell_type' as categorical
... storing 'source' as categorical
... storing 'cell_type' as categorical
... storing 'source' as categorical
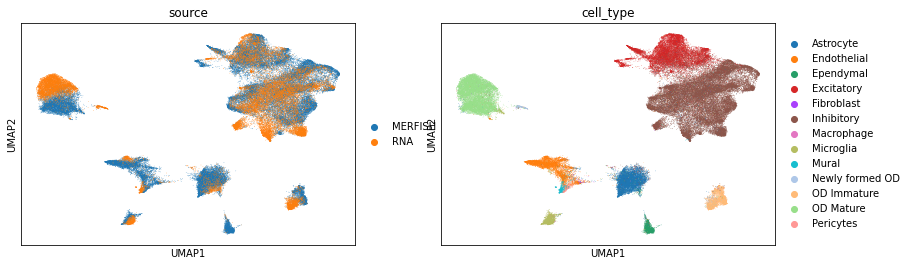
Visualize the data using UMAP according to their cell types and sources.
[10]:
adata_cm_copy = adata_cm.copy()
sc.pp.pca(adata_cm_copy)
sc.pp.neighbors(adata_cm_copy)
sc.tl.umap(adata_cm_copy, min_dist=0.1)
sc.pl.umap(adata_cm_copy, color=['source', 'cell_type'])
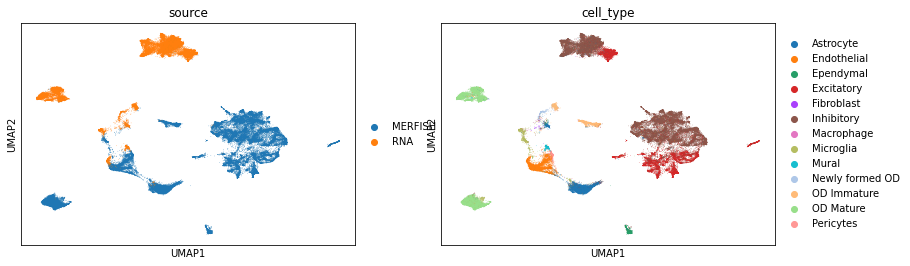
Integrate the MERFISH and scRNA-seq data using both common and dataset-specific genes by Run() function in uniport. The latent representations of data are stored in adata.obs['latent'].
MERFISH and scRNA data integration
[11]:
adata = up.Run(adatas=[adata_merfish, adata_rna], adata_cm=adata_cm, lambda_kl=5.0)
Dataset 0: MERFISH
AnnData object with n_obs × n_vars = 64373 × 155
obs: 'cell_type', 'domain_id', 'source'
var: 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'log1p', 'hvg'
Dataset 1: RNA
AnnData object with n_obs × n_vars = 30370 × 2000
obs: 'cell_type', 'domain_id', 'source'
var: 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'log1p', 'hvg'
Reference dataset is dataset 1
Data with common HVG
AnnData object with n_obs × n_vars = 94743 × 153
obs: 'cell_type', 'domain_id', 'source'
var: 'highly_variable', 'means', 'dispersions', 'dispersions_norm', 'highly_variable_nbatches', 'highly_variable_intersection'
uns: 'log1p', 'hvg'
Epochs: 100%|████████████████████████████████████| 82/82 [19:21<00:00, 14.16s/it, recloss=795.64,klloss=29.19,otloss=4.08]
[12]:
sc.pp.neighbors(adata, use_rep='latent')
sc.tl.umap(adata, min_dist=0.1)
sc.pl.umap(adata, color=['source', 'cell_type'])